| URLs in this document have been updated. Links enclosed in {curly brackets} have been changed. If a replacement link was located, the new URL was added and the link is active; if a new site could not be identified, the broken link was removed. |
Life Science Data Repositories in the Publications of Scientists and Librarians
Biosciences Librarian
University of Minnesota Libraries
Minneapolis, Minnesota
kirle001@umn.edu
Abstract
Bibliographic analysis of the sciences literature indicates that several data repositories are used by science practitioners in their research publications, conference presentations and patents. These specialized resources offer data storage, search, visualization, and sharing capabilities to the science communities of which they are a part. Some science librarians also use data repositories in their publications and in the performance of their professional duties. As the need for established data archives continues to grow, both existing and future data repositories present potential opportunities for the data-related work of science librarians.
Introduction
There are a large number of established science data repositories, data collections and data centers in existence today, many of them readily accessible online. Their scope and community focus is typically quite specialized: ecological studies, oceanographic expeditions, gene sequences, protein structures, toxicological assays, etc. Sponsorship for these resources is diverse and includes federal government agencies, academic and non-profit organizations, individual academic research groups, and the private sector.
In a similar way to the scholarly journal literature and professional conference presentations, data repositories are used by scientists to support aspects of their research, product development and educational activities. Because of their specialized content, the existence of many of these valuable resources is often known only to the scientists with an interest in that specialty, and to the computer technologists, science librarians, and curators/archivists who support those scientists. This context suggests that it continues to be helpful for science librarians to be aware of the content and usage of relevant data resources in their subject areas of responsibility, in order to better inform the faculty, staff, students and professional colleagues that they support.
The overall goal of this study was to explore the usage patterns of life science data repositories in publications by both scientists and librarians. More specifically, the study's research questions were:
- What content and services do life science data repositories provide?
- Do scientists mention life science data repositories in their journal and review articles?
- Do scientists mention life science data repositories in their conference publications?
- Are life science data repositories mentioned in patents?
- Do library professionals mention life science data repositories in their journal and review articles?
- Do library professionals mention life science data repositories in their conference publications?
- What are the relative levels of these usage patterns?
Answers to these questions can help to inform discussions within the library and information science community about future user services development, the continuing educational needs of science librarians, and the complex issues of organizational data stewardship and data curation.
Literature Review
Science, by definition, is a data gathering, experiment- or study-focused activity. Reported studies of the scholarly information gathering and usage behavior of scientists have provided information about how they access and use the professional and research literature (Davis 2004; Haines et al. 2010; Niu et al. 2010). There has also been some work examining their use of data repositories. A study of molecular biology graduate students (Brown 2005) found that they used bioinformatics resources extensively, although they learned about these resources from laboratory colleagues rather than at the library. A 2003 bibliographic analysis study using SciFinder Scholar showed increased use of the GenBank data repository in the science journal literature at that time (Brown 2003).
Reflecting a related trend, a recent web site survey study of local institutional repositories at the top 11 biology graduate programs in the United States uncovered 93 biology-related repositories, suggesting the continuing importance of data archiving and sharing to the life sciences (Brown & Abbas 2010). Another recent web site survey (Marcial & Hemminger 2010) of 100 science data repositories (including several health, biological, and environmental science repositories) also discussed the growth in the number and importance of these resources. This study focused on the general structural characteristics of all types of data repositories, including data deposition and access, metadata, grant and contract support, sponsorship, and preservation policies, and used these observations to provide recommendations for the development and sustainability of science data repositories.
Data repositories are used by some science librarians in their work duties. A number of librarians have discussed their use of some of the National Center for Biotechnology Information's (NCBI) data resources for teaching and training purposes (Dinkelman 2007; Lyon et al. 2006b; MacMillan 2010; Osterbur et al. 2006; Tennant 2005). These teaching opportunities have included library training workshops, course integrated instruction, class lectures, seminar presentations and faculty meetings, and involvement in the coordination of the NCBI's own training programs. Some librarians reported strong support from the science faculty, who realized that a working knowledge of these types of data collections is important for both undergraduate and graduate level life science students. These resources have been used in reference services and research consultations (Lyon et al. 2006b; Osterbur et al. 2006), and to facilitate collaborations between science librarians and non-library bioinformatics support units (Lyon et al. 2006b). This collection of reported work illustrates the diverse and situation-specific strategies that can be used to build library services based on data repositories.
Because many science data repositories are specialized and fairly complex, preparatory training for librarians can be an issue. In addition to the subject-based academic degrees which many science librarians have, available training and continuing education opportunities for librarians in the area of bioinformatics data resources have been discussed in the literature (Alpi 2003; Dinkelman 2007; Tennant 2005). In the past, NCBI has offered numerous in-person training opportunities for both librarians and scientists, such as classes entitled Introduction to Molecular Biology and Information Resources, Advanced Workshop for Bioinformatics Information Specialists, the NCBI Field Guide, and an array of mini courses on several specialized topics. Currently NCBI offers a series of two-day in-person Discovery Workshops in four different areas (Sequences, Genomes and Maps; Proteins, Domains and Structures; NCBI BLAST Services; Human Variation and Disease Genes) and a number of web and video tutorials. All this educational content is accessible at the NCBI education web site (National Center for Biotechnology Education... [cited 2010]). Some librarians (Lyon et al. 2006a; Tennant & Lyon 2007) have also written papers describing the use of the Protein Data Bank and the NCBI's Entrez Gene search interface, the Molecular Modeling Database (also called Structure) and the Cn3D macromolecular visualization and analysis tool.
Because of rapid advances in computer and data sensor technology, the current pace of science data collection is unprecedented, resulting in a phenomenon that some have referred to as the "data deluge" (Borgman et al. 2007; Taming the Data Deluge... [cited 2010]). Some academic libraries and government and other non-profit organizations have been exploring the complexity and necessity of science data archiving (Choudhury 2008; Gabridge 2009; Greenberg 2009; Morris 2010; Ogburn 2010; Witt 2008). One example of an ongoing research project in this area is the Data Conservancy at Johns Hopkins University's Sheridan Libraries, a multiple partner, five-year National Science Foundation (NSF) DataNet project developing new infrastructure for interdisciplinary data curation and preservation, and exploring new data-focused service and collection management roles for research libraries (Hanisch & Choudhury 2009). Thus data archiving is currently an area of active inquiry and ongoing development, and the issue is of interest to science librarians because of the inevitable questions that arise regarding future technological data infrastructure and data support services. As some librarians have already experienced a positive return on their individual investments in data repository experience, a deepened understanding of the current existence and use of science data repositories has the potential to contribute to the discussion and planning of future data support initiatives.
Methods
This bibliographic analysis used metrics (numbers of publications, publication years) for selected life science data repositories, collected from scholarly literature database search tools. Manual data repository web site surveys were also used to collect information about available user services.
Data Repositories Used for the Study: As indicated by the compilation of more than 1,200 databases in the 2010 life science database issue for the publication Nucleic Acids Research (Cochrane & Galperin 2010; Nucleic Acids Research... [cited 2010]), many of these types of resources have been created for the life sciences. To keep the size of this study manageable, 21 repositories (see Tables 1 and 2) were chosen for the study based on one or more of the following general guiding principles:
- Repositories representative of the research and educational interests of life scientists
- Repositories mentioned in the "Instructions to Authors" sections of prominent science journals
- Likely interest for life science librarians based on informal observations of some online science library research guides
- Recently developed repositories reflecting new cutting-edge research objectives in the molecular life sciences
- Repositories with user data deposition features
- Resources with which the author has some working familiarity
- Repositories with free web access, open to the public
Table 1: Life Science Data Repositories - Content
| Repository Name | Start Date | Primary Data Content | Number of Records |
|---|---|---|---|
Chemical Effects in Biological Systems (CEBS) |
2003 |
Toxicogenomics data |
27 studies |
n/a |
Dictionary of small chemical compounds |
597920 entries |
|
Comparative Toxicogenomics Database (CTD) |
2003 |
Chemical-disease and gene-disease relationships |
246514 chemical-gene interactions |
n/a |
single nucleotide polymorphisms (SNPs) |
more than 23000000 SNPs in 58 organisms |
|
Dryad |
n/a |
evolutionary biology, ecology |
292 items |
Ensembl |
2000 |
genome data |
more than 51 species |
GenBank (NCBI) |
1982 |
nucleotide sequences |
more than 122941883 sequences |
2000 |
microarray and high throughput gene expression data |
472195 samples |
|
HapMap |
2005 |
genetic variants (alleles) for four human populations |
more than 3.1 milion SNPs |
Human Metabolome Database (HMDB) |
2007 |
biochemical and clinical data for small molecule metabolites |
8428 compounds |
Mid-1990s |
taxonomic information for plants, animals, fungi, microbes |
490032 |
|
Kyoto Encyclopedia of Genes and Genomes (KEGG) |
1995 |
metabolic pathways, genomes, genes, disease and drug information |
115504 pathways, 1398 organisms |
miRBase |
2003 |
annotated micro RNA sequences |
more than 14000 entries from 133 species |
{National Biological Information Infrastructure (NBII)} |
1993 |
wildlife biology and ecosystems (data collections from multiple organizations) |
n/a |
1988 |
a substantial repository of numerous life science data collections |
n/a |
|
National Center for Ecological Analysis and Synthesis (NCEAS) |
n/a |
ecological studies |
253 |
Protein Data Bank (PDB) |
1976 |
3D structure data for macromolecules (proteins and nucleic acids) |
67322 entries |
PubChem (NCBI) |
2004 |
bioactivity data, medical and toxicological information for small molecules |
28791569 chemical compounds |
2000 |
non-redundant DNA, RNA and protein sequences |
2160929 genomic accessions, |
|
1994 (for prototype) |
phylogenetic trees |
n/a |
|
University of California Santa Cruz Genome Browser (UCSC GB) |
2002 |
genome data |
51 species |
Table Notes:
|
|||
Table 2: Life Science Data Repositories - User Support Services
| Repository Name | Visualization tools | Data analysis tools | Data search tools | Data deposition | Data downloads | Web 2.0 features | Online help |
|---|---|---|---|---|---|---|---|
Chemical Effects in Biological Systems (CEBS) |
Yes |
No |
Yes |
Yes* |
Yes |
No |
No |
No |
No |
Yes |
Yes |
Yes |
No |
Yes |
|
Comparative Toxicogenomics Database (CTD) |
Yes |
No |
Yes |
Yes* |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
|
Dryad |
No |
No |
Yes |
Yes |
Yes |
Yes |
No |
Ensembl |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
GenBank (NCBI) |
Yes |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
|
HapMap |
Yes |
Yes |
Yes |
No |
Yes |
No |
Yes |
Human Metabalome Database (HMDB) |
Yes |
No |
Yes |
No |
Yes |
No |
No |
No |
No |
Yes |
Yes* |
Yes |
No |
No |
|
Kyoto Encyclopedia of Genes and Genomes (KEGG) |
Yes |
Yes |
Yes |
No |
Yes |
No |
Yes |
miRBase |
Yes |
No |
Yes |
Yes |
Yes |
No |
Yes |
{National Biological Information Infrastructure (NBII)} |
Yes |
No |
Yes |
Yes* |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
|
National Center for Ecological Analysis and Synthesis (NCEAS) |
No |
No |
Yes |
Yes |
Yes |
No |
No |
Protein Data Bank (PDB) |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
PubChem (NCBI) |
Yes |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
No |
Yes |
|
Yes |
No |
Yes |
Yes |
Yes |
No |
Yes |
|
University of California Santa Cruz Genome Browser (UCSC GB) |
Yes |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
Table Notes:
|
|||||||
Data Repository Web Site Surveys: Each data repository web site was manually reviewed for content describing its scope, year of establishment, descriptive statistics, available data visualization, analysis and search features, and any Web 2.0 community support features such as blogs, wikis, and RSS feeds.
Journal Article Bibliographic Searches: The scientist journal and review article counts were obtained using two different literature databases, Science Citation Index (SCI in the Web of Science) and PubMed. Librarian and information scientist metrics for journal and review articles were obtained using Library Information Science & Technology Abstracts (LISTA). All the database searches were designed to screen the article titles, abstracts, subject descriptors, and keywords during the time period 1976 to late summer 2010, but did not access the full text of the articles. In additional to total article counts, publication year trends were also obtained from the SCI. For the purpose of contextual comparisons, an estimate of the total number of articles (TNA) referring to biological concepts was obtained using a similar PubMed search strategy for the word stem "biolog."
Conference Publication Searches: The scientist conference article metrics were obtained using SCI with the appropriate document type search filters set to locate publications from meetings and proceedings. The librarian conference article data was collected by manually searching official programs, available at the individual annual conference web sites of three librarian organizations relevant to the life sciences, for relevant presentations and posters during the time period 2007-2010:
- Biomedical & Life Sciences Division, Special Libraries Association (SLA-DBIO)
- Medical Library Association (MLA)
- Science & Technology Section, Association of Colleges and Research Libraries, American Library Association (ACRL-STS)
Patent Searches: The full text of all post-1976 United States patents were searched for the time period 1976 to late summer 2010, using the patent quick search and advanced search tools available at the USPTO web site. Patents prior to 1976 were not searched since the full text of these patents was not directly accessible to the search tools, and information from patents from that time period was not relevant to this study. These searches collected total patent counts, as well as counts filtered by patent sections. The sections selected were title, abstract, description, claims and other references. Similar to the previously described procedure for estimating the PubMed TNA, an estimate of the total number of patents (TNP) referring to biological concepts was obtained by searching for the word stem "biolog."
Results
The results of the web site survey presented in Tables 1 and 2 demonstrate that most of the repositories intentionally scope their content to meet the needs of a specific audience of scientists, and build value-added features that address some of those needs. In addition to data storage and archiving, many repositories provide additional value-added features such as data analysis, visualization tools and data sharing features. Many provide workflows for users to upload their data to the repository. Visualization tools for communicating complex content such as protein three-dimensional structures, genome and gene features, and biological pathways, are provided by several of the repositories and are likely to be considered essential tools by many of the users of these repositories. Some of the repositories are enhanced by community features such as RSS feeds and comment forms. The web site survey also revealed that the TreeBASE and Dryad data repositories have established data archiving partnerships with specific journals.
Figure 1 shows total usage by scientists in their journal articles and reviews of the five most used data repositories; figure 2 shows the same information for the rest of the 21 repositories studied. The total usage data has been split between two figures to accommodate the different vertical axis scaling required to display the usage levels clearly. In the two figures, a total of 350 articles is the cutoff point between moderate usage and higher usage selected to best display all the available usage data. The terminology "usage" in the context of this study is used to specifically refer only to the occurrence of the repository name in the bibliographic metadata, not to the numbers of visitors to the repository web sites or other possible applications of the term. Based on the data in figures 1 and 2, the observed repository usage patterns are fairly similar for the SCI and PubMed literature databases used to collect the data, and the use of more than one literature database in the study results in a more balanced perception of the usage of each data repository because of the somewhat different journal coverage associated with each database.
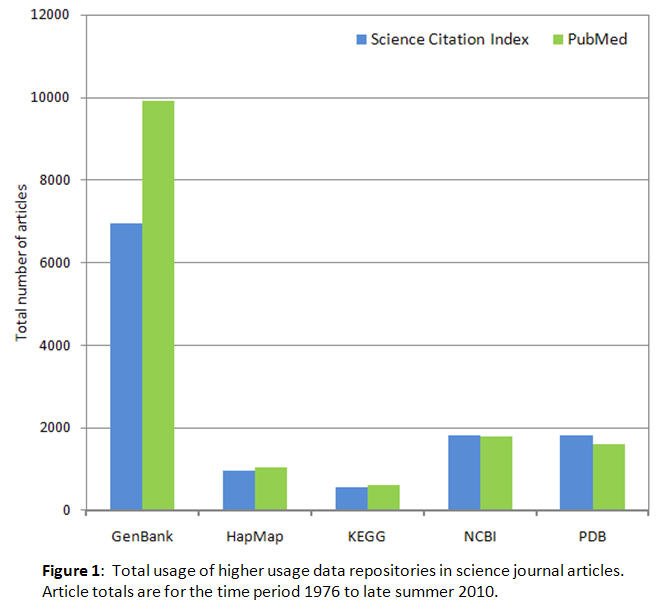
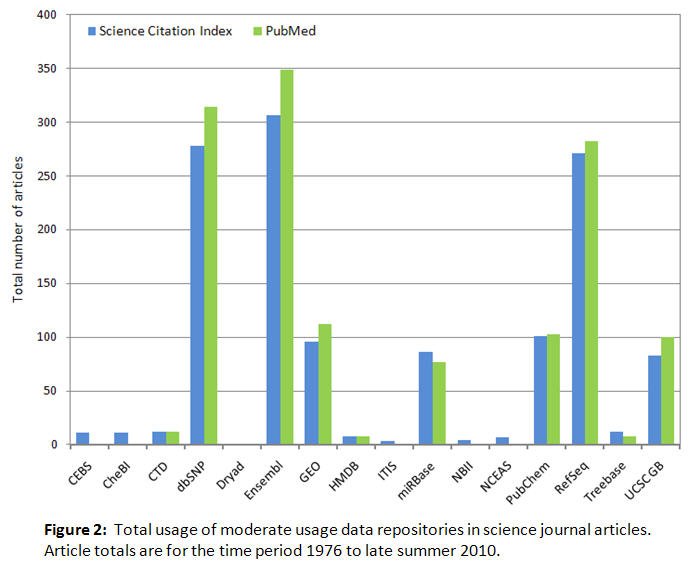
The PubMed TNA, previously defined in the methods section, was 653,371 articles. Thus for example GenBank, found in 9,913 PubMed articles, was 1.5% of the PubMed TNA, and Ensembl, found in 349 PubMed articles, was < 0.1% of the PubMed TNA. The total number of PubMed journal articles for all 21 repositories was 16,337, corresponding to 2.5% of the PubMed TNA.
As for Figures 1 and 2, the annual usage data in Figures 3 and 4 has also been split between two figures to accommodate the different vertical axis scaling required for data display purposes. These two figures show that annual usage levels for the majority of the repositories have grown much more than 100% since their inception. Repositories established a few decades ago, such as the PDB and Genbank, experienced noticeable growth in annual usage beginning in the early 1990s, and now have well established user bases. For example, the PDB annual usage grew from 12 articles per year in 1991 to 133 articles per year in 2009, an increase of 1,108%. The pattern for Genbank is also consistent with earlier trends observed for Genbank (Brown 2003). A number of repositories experienced annual usage growth near the beginning of the last decade and are still increasing. Examples of this are HapMap's 5,900% increase from 4 articles per year in 2003 to 236 articles per year in 2009, the Kyoto Encyclopedia of Genes and Genomes' 2,780% increase from five articles per year in 2000 to 139 articles per year in 2009, and the UCSC Genome Browser's 5,900% increase from two articles per year in 2003 to 19 articles per year in 2009. The NCBI-sponsored repositories (see Tables 1 and 2) also fit this pattern. Ensembl is experiencing annual usage growth again after some reduction in usage in mid-decade. Some recently established repositories such as PubChem and miRBase have moderate levels of usage in publications (20 and 33 articles per year respectively), but currently appear to be increasing. Annual usage for seven other repositories not displayed in Figures 3 and 4 (CEBS, CheBI, Dryad, HMDB, ITIS, NBII, NCEAS) has remained below four articles per year, and clear chronological trends are not yet obvious for these repositories.
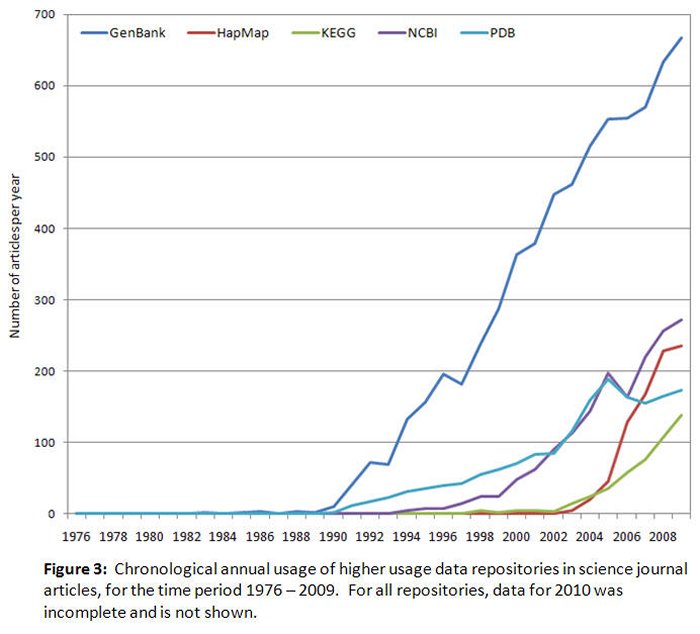
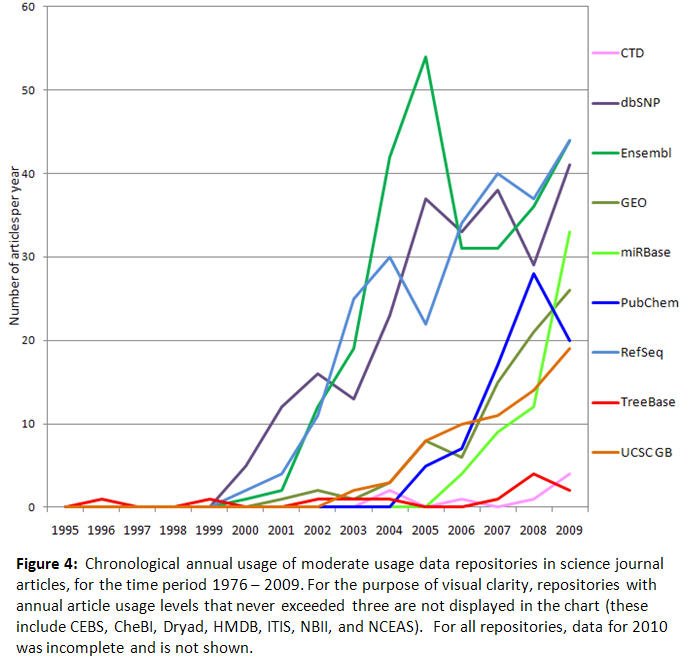
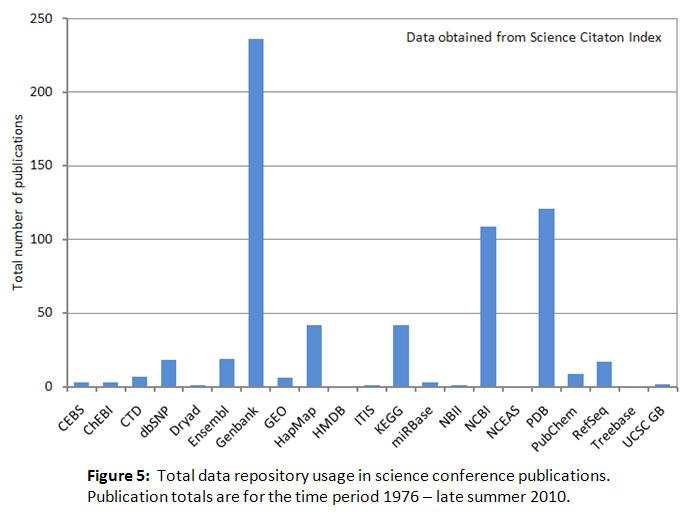
Figure 5 shows that total conference publication usage by scientists of some data repositories ranges from 236 publications for Genbank to below 50 publications for 15 other repositories , and zero publications for three repositories. Thus overall the observed total usage levels are lower than for journal publications. This possibly reflects the difficulty of comprehensively indexing these types of publications in literature databases, and the fewer available venues for presentation than for publication.
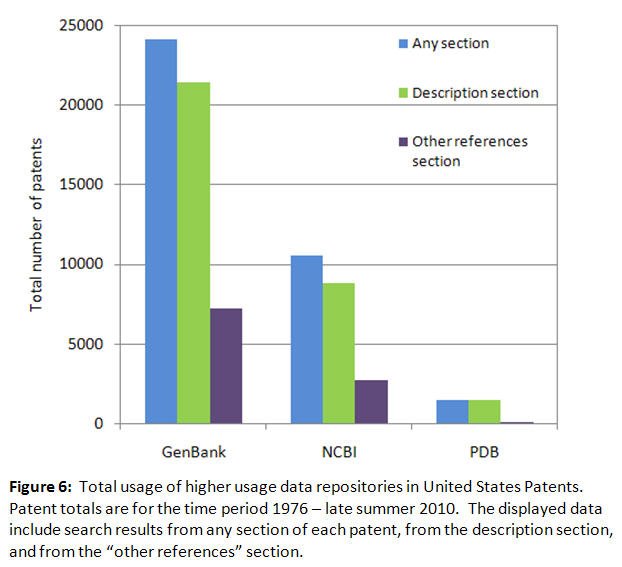
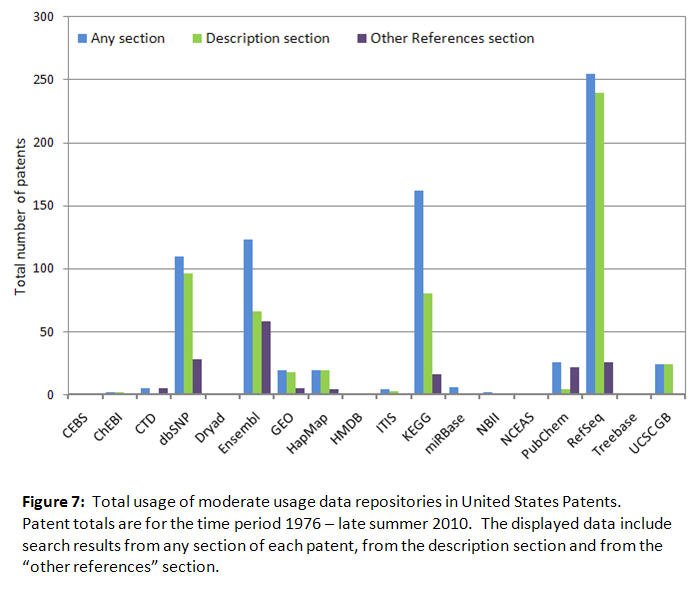
As with Figures 1-4, the total usage data for patents has been split between Figures 6 and 7 to accommodate the different vertical axis scaling required to display the higher and moderate usage levels clearly, with a total of 500 patents selected as the cutoff point between the two levels. This data shows that total usage in all patent sections for higher usage repositories such as Genbank (24,110 patents) and the PDB (1,511 patents) is greater than moderate usage repositories such as RefSeq (255 patents) and dbSNP (110 patents). Patents are not yet using some of the most recently established repositories such as CEBS and HMDB. The total number of patents for all 21 repositories was 36,907, 14.7% of the observed TNP of 251,595 for all patent sections, indicating that these resources are sometimes considered relevant for the establishment of intellectual property rights. The figures also document the notable usage of repository names in the description and "other references" sections of the patents. Observed usage was much less in the claims, abstract and title sections of the patents (data not included).
Librarians use some data repositories in their journal and review articles, but the total usage levels are lower than that of the science community. Based on LISTA searches, the most frequently used repositories were NCBI (41 articles), Genbank (22 articles), NBII (17 articles), PDB (8 articles) and Pubchem (7 articles). Many other repository names were not found in the library science journal literature. LISTA searches did not reveal usage of any repositories except Genbank in librarians' conference presentations. Based on a manual review of the archived official MLA annual conference programs, the number of presentations or posters mentioning the NCBI ranged from two to six per year during the time period 2007-2010, and one presentation in this time period also mentioned the UCSC genome browser. Similarly, the number of presentations for ALA STS mentioning the NCBI ranged from zero to one per year. SLA DBIO had the greatest variety of repositories mentioned (NCBI, Protein Data Bank, Genbank, Pubchem, etc.), and ranged from zero to three presentations per year. Collectively this information indicates that data repositories remain a specialized but sustained interest for science librarians.
Discussion and Conclusions
Several life science data repositories provide important support for the work of science practitioners, and their annual usage in journal articles continues to increase. In addition, the repository web site capabilities are significant, providing value-added data analysis and visualization tools, specialized search capabilities, and user data download and upload workflows. Percentage comparisons of scientists' total usage of the 21 data repositories included in this study with estimates of the total number of publications referring to biological concepts ranged from 2.5% for journal articles to 14.7% for patents, reflecting the specialized nature of these resources.
Usage levels of these repositories by librarians in their own scholarly products, although less than those of the science community are still notable. The differences in usage levels reflect, in part, the large differences in the numbers of life scientists vs. librarians; 2008 estimates of the sizes of the two professions were 156,000 for biological scientists (Occupational Outlook Handbook... [cited 2010]) and 27,030 for academic librarians (Number Employed in Libraries ... [updated 2010]). In the case of both the librarian and scientist usage data, it should be kept in mind that this measurable usage is only a subset of all the likely usage, since most users accessing a data repository do not immediately produce a paper or presentation as a result of that access, and many may never do so. Also, the literature database search tools for this study did not directly access the full text of the documents. Since the bibliographic analysis constraints were similar when using the literature databases, relative comparisons are still possible.
The clear growth of many science data repositories presents both opportunities and challenges for libraries. As previously described, it has been demonstrated that science librarians with a working familiarity with these types of resources have an effective additional skill with which to successfully provide reference and instructional services to the scientists and students that they support, and there are educational resources available to assist librarians with acquiring these capabilities. The development of these skill sets can help to minimize the risk of science libraries being bypassed in this important area of growth for scientists; it is recognized that building and maintaining these skills can be an ongoing challenge for the multitasking librarian. Bibliographic analysis and bibliometric studies of data repository usage patterns can help to provide near-term guidance for areas of repository growth and the relative impact of various repositories, and thus assist in the choices that librarians make regarding where to invest limited time and staff resources.
Looking beyond the existing library services already described, data repository usage by scientists presents possible opportunities for librarians. Several repositories play a critical role in the editorial policies of many life science journals. Prominent science publications such as Nature (Availability of data... [No date]), Science Magazine (General Information for Authors... [cited 2010]), Journal of Biological Chemistry (The Journal of Biological Chemistry... [updated 2010]), Molecular and Cellular Biology (Instructions to Authors... [cited 2010]), PLoS Biology (PLoS Biology... [cited 2010]), BMC Biology (Instructions for BMC Biology authors ... [updated 2010]) and BMC Bioinformatics (Instructions for BMC Bioinformatics authors ... [updated 2010]), require authors submitting manuscripts to upload supporting experimental data to specific data repositories relevant to the subject of the manuscript, when these are available. The accession numbers assigned to the datasets by the repositories become part of the final journal publication. These journal editorial policies contribute to the influx of data into these repositories, and the timing of data submission fits into the natural workflow of science practitioners, since they have direct access to all the data files during the manuscript preparation process. These data repositories also have flexible policies that do not require that data be part of a formal journal publication in order to be submitted, facilitating submissions beyond the scope of the journal article publication process. The existence of this direct connection between research article publication and data repositories has the potential to create some opportunities for science librarians to engage faculty, students and staff who need guidance and consultation support in locating, selecting and using repositories for data preservation that are appropriate for the publication process.
Data repositories and archives play an important role in data management and sharing. The National Institutes of Health (NIH) currently requires that researchers who receive its grant funding have a data sharing plan in place, which can include the use of a data archive (NIH Data Sharing Policy ... [updated 2007]). The National Science Foundation (NSF) has recently announced that it will begin requiring data management plans in new grant proposals in mid-January 2011 (Dissemination and Sharing... [updated 2010]). Components of the required NSF data management plan include descriptions of the types of data to be produced by the research, data and metadata standards, policies for data access and sharing, and planned strategies for data preservation and sharing. One possible alternative for long-term preservation is the deposition of research data in an existing public data repository (Donnelly & Jones 2009). In a similar way to the journal data policies already described, these grant funding agency initiatives also create potential opportunities for science librarians to provide data support services for the clients they support (Delserone 2008; Garritano & Carlson 2009; Witt 2008; Data Management... [cited 2010]).
When librarians are familiar with existing subject-based data repositories, they can provide data support services by guiding researchers to relevant data archives for their data management plans. For some areas of study, there is no well-known data repository for the research data. Although many scientists will already be familiar with the data repository appropriate for their datasets, others may require significant assistance with exploring possible alternatives such as subject-based and general purpose-repositories, local institutional repositories and local data storage options. Some examples of data repository initiatives with library science sponsors include Cornell University's Datastar and the University of North Carolina's Dryad (Greenberg 2009). The latter is recommended by the journal Nature (Availability of data... [No date]) as one of the repositories for biology datasets. Collectively these types of projects demonstrate the growing potential of the involvement of academic research libraries in science data services and support.
The annual usage in the sciences literature of many of the data repositories described in this study is trending upwards over time even though these resources are primarily of interest only to specialized communities of scientists. This pattern of sustained growth indicates that these data repositories will continue to play an important role in the sciences at least for the near term. The academic research library community is increasing its interest and investment in science data repositories, archiving, curation and related services, and some science research funding agencies and journal publishers are increasing their data archiving requirements. This overall situational context suggests that these types of resources will contribute to the suite of support services that science librarians will offer their clients in the future. Bibliographic analysis, such as the type described in this study, is a possible strategy that can be used by science librarians to monitor data repository growth trends, and an awareness of these trends is one factor that can be of assistance in the determination of the level of investment, placement and timing of library-based science data support and educational services.
References
About the PDB Archive and the RCSB PDB. [Internet]. [Date unknown.] Research Collaboratory for Structural Bioinformatics. [Cited 2010 Aug 22]. Available from: http://www.pdb.org/pdb/static.do?p=general_information/about_pdb/index.html
Alpi, K. 2003. Bioinformatics training by librarians and for librarians: developing the skills needed to support molecular biology and clinical genetics information instruction. Issues in Science and Technology Librarianship [Internet]. [Cited 2010 Aug 29];37 (Spring). Available from: http://www.istl.org/03-spring/article1.html
Availability of Data and Materials. [Internet]. [No date]. Nature Publishing Group. [Cited 2010 Aug 22]. Available from: http://www.nature.com/authors/editorial_policies/availability.html
Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J. & Sayers, E.W. 2010. GenBank. Nucleic acids research, 38(Database issue):46-51.
Borgman, C., Wallis, J. & Enyedy, N. 2007. Little science confronts the data deluge: habitat ecology, embedded sensor networks, and digital libraries. International Journal on Digital Libraries, 7(1):17-30.
Brown, C. & Abbas, J.M. 2010. Institutional digital repositories for science and technology: a view from the laboratory. Journal of Library Administration, 50(3):181-215.
Brown, C. 2005. Where do molecular biology graduate students find information? Science & Technology Libraries, 25(3):89-104.
Brown, C. 2003. The changing face of scientific discourse: analysis of genomic and proteomic database usage and acceptance. Journal of the American Society for Information Science & Technology, 54(10):926-938.
Choudhury, G.S. 2008. Case study in data curation at Johns Hopkins University. Library Trends, 57(2):211-220.
Cochrane, G.R. & Galperin, M.Y. 2010. The 2010 nucleic acids research database issue and online database collection: a community of data resources. Nucleic Acids Research, 38(suppl 1): D1-D4.
Data Management and Publishing. [Internet]. MIT Libraries. [Cited 2010 Nov 23]. Available from: {https://libraries.mit.edu/data-management/}
Davis, A.P., Murphy, C.G., Saraceni-Richards, C.A., Rosenstein, M.C., Wiegers, T.C. & Mattingly, C.J. 2009. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Research 37(Database issue):786-792.
Davis, P.M. 2004. Information-seeking behavior of chemists: A transaction log analysis of referral URLs. Journal of the American Society for Information Science and Technology, 55(4):326-332.
Delserone, L.M. 2008. At the watershed: preparing for research data management and stewardship at the University of Minnesota Libraries. Library Trends 57( 2): p202-210.
Dinkelman, A.L. 2007. "See a need, fill a need" -- reaching out to the bioinformatics research community at Iowa State University. Issues in Science & Technology Librarianship(52) [Internet]. [Cited 14 Mar 2011]. Available from: http://www.istl.org/07-fall/refereed1.html
Dissemination and Sharing of Research Results. [Internet]. [Updated 2010 Nov 16]. National Science Foundation [Cited 2010 Nov 20]. Available from: http://www.nsf.gov/bfa/dias/policy/dmp.jsp
Donnelly, M. & Jones, S. 2009. Data Management Plan Content Checklist: Draft Template for Consultation. [Internet]. Data Curation Centre. Available from: http://www.dcc.ac.uk/sites/default/files/documents/templates/DMP_checklist.pdf
Flicek, P., Aken, B.L., Ballester, B., Beal, K., Bragin, E., Brent, S., Chen, Y., Clapham, P., Coates, G., Fairley, S., et al. 2010. Ensembl's 10th year. Nucleic Acids Research, 38 (Database issue):557-562.
Gabridge, T. 2009. The last mile: liaison roles in curating science and engineering research data. Research Library Issues (265):15-21.
Garritano, J.R. & Carlson, J.R. 2009. A subject librarian's guide to collaborating on e-science projects. Issues in Science and Technology Librarianship [Internet]. [Cited 2010 Aug 29];57 (Spring). Available from: http://www.istl.org/09-spring/refereed2.html
General Information for Authors. [Internet]. American Association for the Advancement of Science. [Cited 2010 Aug 25]. Available from: {http://www.sciencemag.org/about/authors/prep/gen_info.dtl}
Greenberg, J. 2009. Theoretical considerations of lifecycle modeling: an analysis of the Dryad Repository demonstrating automatic metadata propagation, inheritance, and value system adoption. Cataloging & Classification Quarterly, 47(3):380-402.
Griffiths-Jones, S., Saini, H.K., van Dongen, S. & Enright, A.J. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Research, 36 (Database issue):154-158.
Haines, L.L., Light, J., O'Malley, D. & Delwiche, F.A. 2010. Information-seeking behavior of basic science researchers: implications for library services. Journal of the Medical Library Association, 98(1):73-81.
Hanisch, R. & Choudhury, S. 2009. The Data Conservancy: Building a Sustainable System for Interdisciplinary Scientific Data Curation and Preservation. [Internet]. [Updated 2009]. JScholarship [Cited 2010 Nov 20]. Available from http://jhir.library.jhu.edu/handle/1774.2/34018
Instructions for BMC Bioinformatics authors. [Internet]. [Updated 2010 Aug 9]. BioMed Central Ltd. [Cited 2010 Aug 22]. Available from: http://www.biomedcentral.com/bmcbioinformatics/ifora/
Instructions for BMC Biology authors. [Internet]. [Updated 2010 Aug 9]. BioMed Central Ltd. [Cited 2010 August 22]. Available from: http://www.biomedcentral.com/bmcbiol/ifora/
Instructions to Authors: Molecular and Cellular Biology, Editorial Policy. [Internet]. American Society for Microbiology. [Cited 2010 Aug 22]. Available from: {http://journals.asm.org/site/misc/authors.xhtml}
International HapMap Consortium, Frazer, K.A., Ballinger, D.G., Cox, D.R., Hinds, D.A., Stuve, L.L., Gibbs, R.A., Belmont, J.W., Boudreau, A., Hardenbol, et al. 2007. A Second Generation Human Haplotype Map of Over 3.1 Million SNPs. Nature, 449(7164):851-861.
The Journal of Biological Chemistry: Important Submission Instructions for Authors. [Internet]. [Updated Aug 19, 2010]. American Society for Biochemistry and Molecular Biology. [Cited 2010 Aug 22]. Available from: http://www.jbc.org/site/misc/ifora.xhtml
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., Katayama, T., Kawashima, S., Okuda, S., Tokimatsu, T. & Yamanishi, Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Research, 36 (Database issue):D480-4.
Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M. & Haussler, D. 2002. The human genome browser at UCSC. Genome Research, 12(6):996-1006.
Lyon, J.A., Tennant, M.R. & Danielson, B. 2006a. Introducing Protein Data Bank, Molecular Modeling Database, and Cn3D. Journal of Electronic Resources in Medical Libraries, 3(3):1-20.
Lyon, J.A., Tennant, M.R., Messner, K.R. & Osterbur, D.L. 2006b. Carving a niche: establishing bioinformatics collaborations. Journal of the Medical Library Association, 94(3):330-335.
MacMillan, D. 2010. Sequencing genetics information: integrating data into information literacy for undergraduate biology students. Issues in Science & Technology Librarianship. [Internet]. [Cited 29 Mar 2011]. Available from: http://www.istl.org/10-spring/refereed3.html
Marcial, L.H. & Hemminger, B.M. 2010. Scientific data repositories on the web: an initial survey. Journal of the American Society for Information Science & Technology, 61(10): 2029-2048.
Morris, S.P. 2010. The North Carolina Geospatial Data Archiving Project: challenges and initial outcomes. Journal of Map & Geography Libraries, 6(1):26-44.
National Center for Biotechnology Education. [Internet]. National Library of Medicine. [Cited 2010 Aug 23]. Available from: http://www.ncbi.nlm.nih.gov/Education/
NIH Data Sharing Policy. [Internet]. [Updated 2007 Apr 17]. Office of Extramural Research. [Cited 2010 Sept 8]. Available from: http://grants.nih.gov/grants/policy/data_sharing/
Number Employed in Libraries: ALA Library Fact Sheet 2. [Internet]. [Updated 2010 Mar]. American Library Association. [Cited 2010 Sept 1]. Available from: http://www.ala.org/ala/professionalresources/libfactsheets/alalibraryfactsheet02.cfm
Niu, X., Hemminger, B.M., Lown, C., Adams, S., Brown, C., Level, A., McLure, M., Powers, A., Tennant, M.R. & Cataldo, T. 2010. National study of information seeking behavior of academic researchers in the United States. Journal of the American Society for Information Science & Technology, 61(5):869-890.
Nucleic Acids Research. [Internet]. Oxford University Press [Cited 2010 Aug 31]. Available from: http://nar.oxfordjournals.org/
Occupational Outlook Handbook, 2010-11 Edition. [Internet]. Bureau of Labor Statistics [Cited 2010 Aug 25]. Available from: (http://www.bls.gov/ooh/}
Ogburn, J.L. 2010. The imperative for data curation. portal: Libraries & the Academy, 10(2):241-246.
Osterbur, D.L., Alpi, K., Canevari, C., Corley, P.M., Devare, M., Gaedeke, N., Jacobs, D.K., Kirlew, P., Ohles, J.A., Vaughan, K.T., Wang, L., Wu, Y. & Geer, R.C. 2006. Vignettes: diverse library staff offering diverse bioinformatics services. Journal of the Medical Library Association, 94(3):306-306.
PLoS Biology Editorial and Publishing Policies. [Internet]. PLoS.org [Cited 2010 Aug 22]. Available from: {http://journals.plos.org/plosbiology/s/materials-and-software-sharing}
Tennant, M.R. 2005. Bioinformatics librarian: meeting the information needs of genetics and bioinformatics researchers. Reference Services Review, 33(1):12-19.
Tennant, M.R. & Lyon, J.A. 2007. Entrez Gene: a gene-centered "information hub". Journal of Electronic Resources in Medical Libraries, 4(3):53-78.
Taming the Data Deluge: DICE Center established at UNC at Chapel Hill. [Internet]. UNC SILS. [Cited 2010 Aug 31]. Available from: http://sils.unc.edu/news/2009/DiceCenter
Wang, Y., Bolton, E., Dracheva, S., Karapetyan, K., Shoemaker, B.A., Suzek, T.O., Wang, J., Xiao, J., Zhang, J. & Bryant, S.H. 2010. An overview of the PubChem BioAssay resource. Nucleic Acids Research, 38 (Database issue):255-266.
Waters, M., Stasiewicz, S., Merrick, B.A., Tomer, K., Bushel, P., Paules, R., Stegman, N., Nehls, G., Yost, K.J., Johnson, C.H., et al. 2008. CEBS - Chemical Effects in Biological Systems: a public data repository integrating study design and toxicity data with microarray and proteomics data. Nucleic Acids Research, 36 (Sp. Iss. SI):892-900.
Wishart, D.S., Knox, C., Guo, A.C., Eisner, R., Young, N., Gautam, B., Hau, D.D., Psychogios, N., Dong, E., Bouatra, S., et al. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research, 37 (Database issue):603-610.
Witt, M. 2008. Institutional repositories and research data curation in a distributed environment. Library Trends, 57(2):191-201.
| Previous | Contents | Next |
